Pathogen Genomics Centers of Excellence
Predicting Salmonella Outbreaks
Outbreaks of dangerous pathogens like Salmonella and E. coli from food products are all too common. For example, the period of 2017-2019 saw thousands of infections of Salmonella Reading, a serotype of the species Salmonella enterica, from ground turkey.
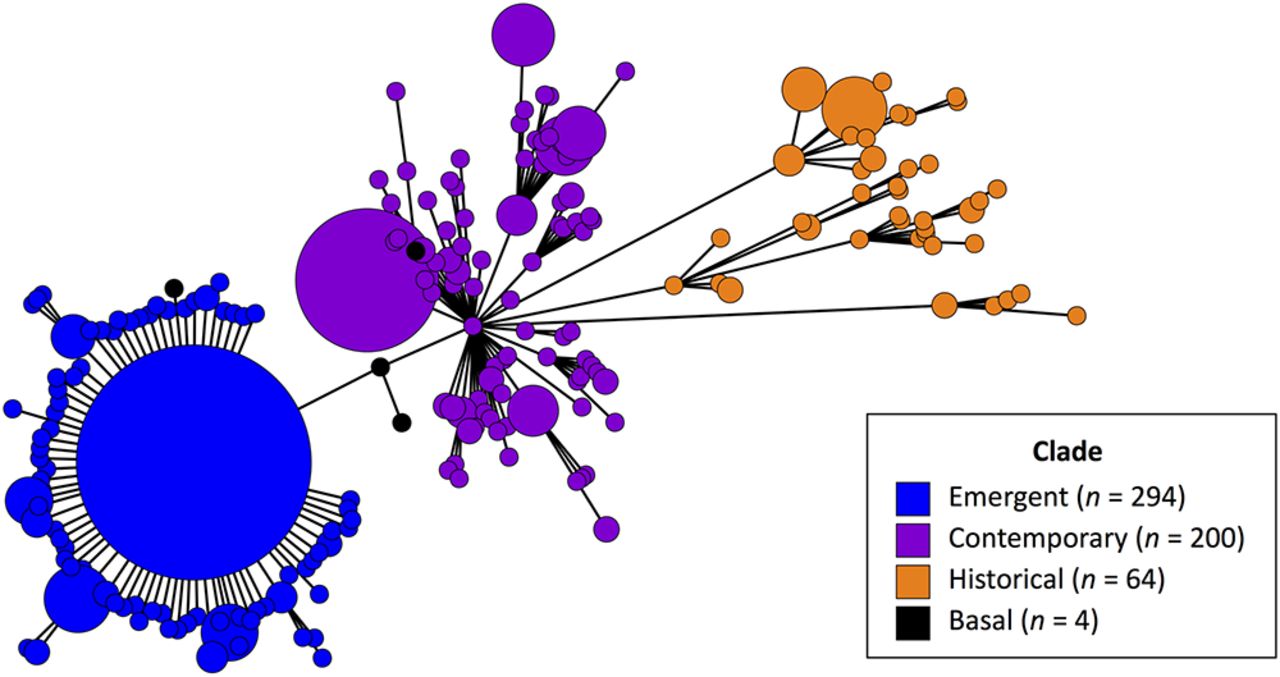
One reason there have been so many outbreaks of Salmonella Reading is that its genetic makeup changed very quickly. As with all Salmonella, efforts to control one particular strain prompts new strains to develop. Since 2016, researchers have found three distinct phylogenetic clades, i.e. steps in evolutionary development, in Salmonella Reading.
When a new strain of a foodborne pathogen arises, medical and public health professionals must respond quickly. The search for the source of an outbreak can typically begin only after a substantial number of people have been affected. If, however, we could predict when outbreaks are likely to occur, food producers could address the problem much earlier. This would greatly reduce the number of people sickened.
Identifying dangerous Salmonella early
Dr. Tim Johnson of the University of Minnesota is leading a project, funded by the Pathogen Genomics Centers of Excellence (PGCoE), to identify high-risk Salmonella strains before they make people sick. The Minnesota Public Health Laboratory is assisting Dr. Johnson with the necessary high-resolution genomic analyses, rapid laboratory screens, and predictive modeling.
The first step is to define the genomes, or complete sets of genes, of the Salmonella that are found in food animal production. There are thousands of serovars, or variations, of Salmonella, so Dr. Johnson and his team have been studying the ones that tend to proliferate in food animals and/or are the most dangerous to humans.
Once the most relevant Salmonella serovars are found, the next step is to identify the strains and gene combinations within them that bring about their “success” in spreading and sickening people. The team then develops tools to surveil the genomes of Salmonella for these strains and gene combinations.
The main result of the project is a simple-to-use, web-based tool which can be used by both public health agencies and food producers for real-time surveillance and early identification of high-risk Salmonella. Food producers will then be able to identify dangerous strains quickly and respond to them on farm before they cause foodborne disease outbreaks. The tool, called Salmonella Predictive Risk Assessment Tool (SPRAT), enables the visualization of real-time Salmonella data extracted from NCBI’s Pathogen Detection platform. As illustrated below with Salmonella Reading, it enables users to identify changes over time in Salmonella populations that may present an enhanced human health risk. This tool will be publicly available in Summer 2024 at sprat.umn.edu.